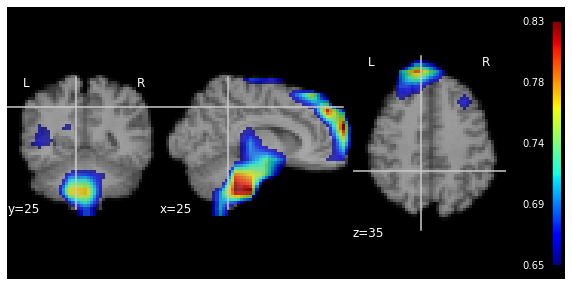
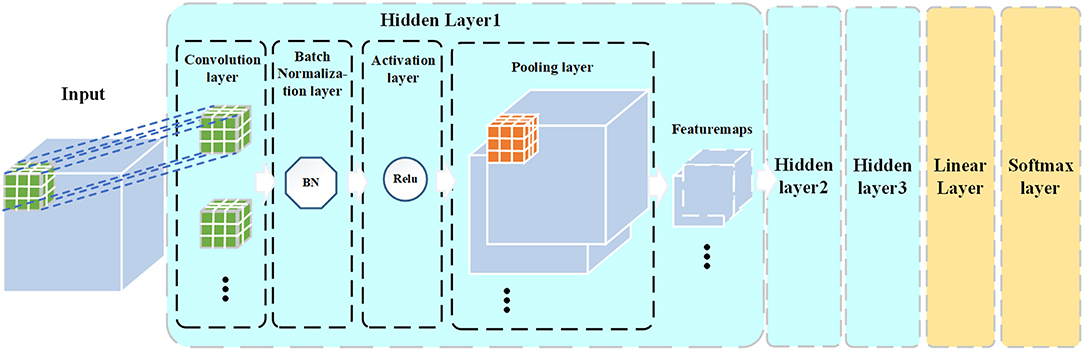
MRI classification with 3D CNN
1. Introduction
In this notebook we will explore simple 3D CNN classificationl model on pytorch from the Frontiers in Neuroscience paper: https://www.frontiersin.org/articles/10.3389/fnins.2019.00185/full. In the current notebook we follow the paper on 3T T1w MRI images from https://www.humanconnectome.org/.
Our goal will be to build a network for MEN and WOMEN brain classification, to explore gender influence on brain structure and find gender-specific biomarkers.
Proceeding with this Notebook you confirm your personal acess to the data. And your agreement on data terms and conditions.
Importing needed libs
Mounting Google Drive to Collab Notebook. You should go with the link and enter your personal authorization code:
Get the data. Add a shortcut to your Google Drive for
labels.npyandtensors.npy.
Shared link: https://drive.google.com/drive/folders/1GCIXnK6ly5l_LADanpLmvtZ6YbqPUamQ?usp=sharing
Let's watch the data. We will use nilearn package for the visualisation:
https://nilearn.github.io/modules/generated/nilearn.plotting.plot_anat.html#nilearn.plotting.plot_anat
|████████████████████████████████| 9.6 MB 28.1 MB/s
Questions:
What is the size of image (file)?
That is the intensity distribution of voxels?
2. Defining training and target samples
From the sourse article:
The original data were too large to train the model and it would cause RESOURCE EXAUSTED problem while training due to the insufficient of GPU memory. The GPU we used in the experiment is NVIDIAN TITAN_XP with 12G memory each. To solve the problem, we scaled the size of FA image to [58 × 70 × 58]. This procedure may lead to a better classification result, since a smaller size of the input image can provide a larger receptive field to the CNN model. In order to perform the image scaling, “dipy” (http://nipy.org/dipy/) was used to read the .nii data of FA. Then “ndimage” in the SciPy (http://www.scipy.org) was used to reduce the size of the data.
3. Defining Data Set
4. Defining the CNN model architecture
At first check if we have GPU onborad:
5. Training the model
Training first 20 epochs:
K-Fold model validation:
Questions:
What is the purpose of K-Fold in that experiment setting?
Can we afford cross-validation in regular DL?
Model save
What else?
MRI classifcation model interpretation
Visit: https://github.com/kondratevakate/InterpretableNeuroDL
Meaningfull perturbations on MEN brains prediction: