Path: blob/main/seminar4/IndividualAnalysis.ipynb
193 views
GLM: 1st-level Analysis
The objective in this example is to model the timecourse of each voxel. This will allow us to answer question regarding to the voxel response to different stimuli.
We will model the activation at each voxel as a weighed sum of explanatory variables and error term
or in matrix notation
which can be solved with Ordinary Least Squares regression.
The parameters represent the contribution of each variable to the voxel activation.
The error terms are assumed to be independent and identically distributed.
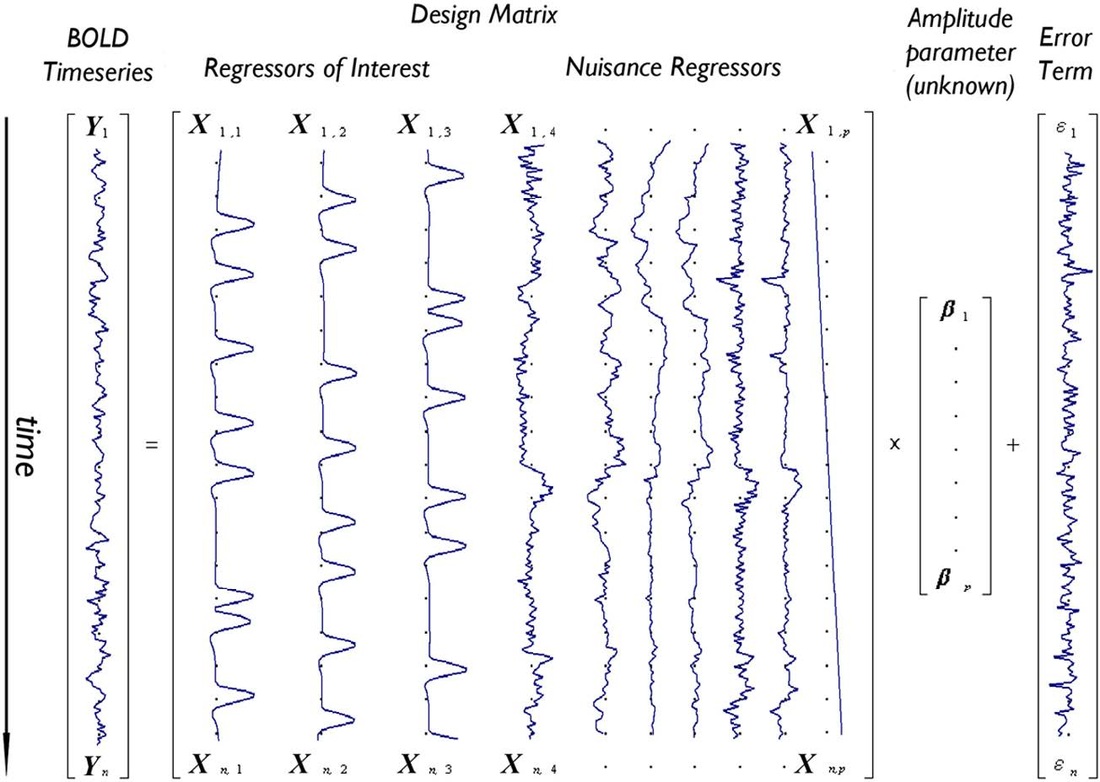
The matrix 𝑋 is also called design matrix.It is up to the researcher which factors to include in the design matrix. The design matrix contains factors that a related to the hypothesis the researcher want to answer. Furthermore, sometimes in it are also included factors that are not related to the hypothesis, but are know sources of variability (nuisance factors).
Now lets take a look how we take into account the delayed bold response. The hemodynamic response function might look something like this:
In the design often we want to include variables, that indicate presence or absence of a stimuli. Let's say we had the 3 stimuli during the timecourse
Next we have to convolve signal with our hrf function
After that we can fit our fMRI data to the design Matrix and use GLM to for hypotesis testing. We can evaluate whether a given factor has a considerable contribution by its coefficient . With t-test we can evaluate whether > 0. We can also test hypothesis of the form > .
The general form of the hypothesis tests which forms a contrast is:
Typical values for are 1 and -1. Typically contrasts are express as:.
For example the contrast [1, 0] tests > 0.
This contrasts form a t-statistics (Recall that coefficient/std(coefficient) in OLS regression follow t-distribution with n-p-1 df).
We can combine several contrasts and form F-statics. In the contexts of fMRI the F-tests help us answer questions like "Is effect A or effect B or effect C, or any combination of them, significantly non-zero?".
Recall that F test is used for comparison between reduced model RF and full model FM:
where the reduced model have k distinct parameters
Combining the test results from all voxels we get statistical map of brain activity. Now that the theory out of our way we can see how it is done in Nipype.
1st Leval Analysis in Nipype
We will use the preprocessed files we got and run 1st-level analysis (individual analysis) for each subject. We will perform the following steps:
Extract onset times of stimuli from TVA file
Specify the model (TR, high pass filter, onset times, etc.)
Specify contrasts to compute
Estimate contrasts
Prepare design matrix
Let's take a look how the stimuli onset and duration look like.This information is store in a tsv file. This file will help us build the design matrix.
The three different conditions in the fingerfootlips task are:
finger
foot
lips
Initiate Nodes
Specify contrasts
We are gona perform several T tests and one F test. Recall the general form of the hypothesis are
Specify input & output stream
Specify where the input data can be found & where and how to save the output data.
Specify Workflow
Create a workflow and connect the interface nodes and the I/O stream to each other.
Visualize the workflow
Run the Workflow
Run the 1st-level analysis workflow.
Run with 'Linear' plugin if multiprocessing frozen during execution
Inspect output
Let's check the structure of the output folder, to see if we have everything we wanted to save. You should one image for each subject and contrast (con_*.nii for T-contrasts and ess_*.nii for F-contrasts)
Visualize results
Let's look at the contrasts of one subject that we've just computed.
Sources:
General Linear Model for Neuroimaging
Special thanks to Michael Notter for the wonderful nipype tutorial