fMRI data preprocessing
Preprocessing
In this workflow we will conduct the following steps:
1. Coregistration of functional images to anatomical images (according to FSL's FEAT pipeline)
Co-registrationis the process of spatial alignment of 2 images. The target image is also called reference volume. The goodness of alignment is evaluated with a cost function.
We have to move the fmri series from fmri native space:
To native anatomical space:
2. Motion correction of functional images with FSL's MCFLIRT
The images are aligned with rigid transformation - rotations, translations, reflections. Then spatial interpolation is done, so as there was no movements.

3. Slice Timing correction
The brain slices are not acquired at the same time. Therefore, interpolation is done between the nearest timepoints 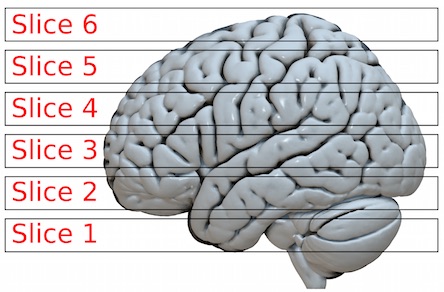
4. Smoothing of coregistered functional images with FWHM set to 5/10 mm
5. Artifact Detection in functional images (to detect outlier volumes)
So, let's start!
Imports
First, let's import all the modules we later will be needing.
Experiment parameters
It's always a good idea to specify all parameters that might change between experiments at the beginning of your script. We will use one functional image for fingerfootlips task for ten subjects.
Specify Nodes for the main workflow
Initiate all the different interfaces (represented as nodes) that you want to use in your workflow.
Coregistration Workflow
Initiate a workflow that coregistrates the functional images to the anatomical image (according to FSL's FEAT pipeline).
Specify input & output stream
Specify where the input data can be found & where and how to save the output data.
Specify Workflow
Create a workflow and connect the interface nodes and the I/O stream to each other.
Visualize the workflow
It always helps to visualize your workflow.
Run the Workflow
Now that everything is ready, we can run the preprocessing workflow. Change n_procs to the number of jobs/cores you want to use. Note that if you're using a Docker container and FLIRT fails to run without any good reason, you might need to change memory settings in the Docker preferences (6 GB should be enough for this workflow).
Run with 'Linear' Plugin, for one process per subject, if multiprocessing failed. Read more: https://nipype.readthedocs.io/en/1.1.0/users/plugins.html
Inspect output
Let's check the structure of the output folder, to see if we have everything we wanted to save.
Visualize results
Let's check the effect of the different smoothing kernels.
Now, let's investigate the motion parameters. How much did the subject move and turn in the scanner?
There seems to be a rather drastic motion around volume 102. Let's check if the outliers detection algorithm was able to pick this up.
Alternative for motion artifacts detection ICA-based Automatic Removal Of Motion Artifact
Dataset: A test-retest fMRI dataset for motor, language and spatial attention functions
Special thanks to Michael Notter for the wonderful nipype tutorial
Example of fmriprep run
Example of running fmriprep
will cause an error, first, we have to create BIDS validated dataset